 |
SMILE
v2.5
Schwarzschild Modelling Interactive expLoratory Environment
|
 |
SMILE
v2.5
Schwarzschild Modelling Interactive expLoratory Environment
|
Ferrers density profile (with finite size R and density (1-(r/R)^2)^2). More...
#include <potential.h>
Public Member Functions | |
| CPotentialFerrers (double _mass, double _R, double _q, double _p) | |
| virtual CPotential * | clone () const |
| Return a pointer to a copy of this instance of potential. More... | |
| virtual POTENTIALTYPE | PotentialType () const |
| enumerable potential type | |
| virtual const char * | PotentialName () const |
| string representation of potential type | |
| virtual SYMMETRYTYPE | symmetry () const |
| returns symmetry type of this potential | |
| virtual double | Rho (double X, double Y, double Z, double t=0) const |
| returns density at given coordinates, this should obviously be overriden in derivative classes | |
| virtual double | Phi (double X, double Y, double Z, double t=0) const |
| Return potential at a given spatial point (possibly a time-varying one). More... | |
| virtual void | Force (const double xyz[N_DIM], const double t, double *force, double *forceDeriv=NULL) const |
| Compute forces and, optionally, force derivatives at a given point. More... | |
| virtual double | getGamma () const |
| returns inner density slope estimate (only used in BSE potential expansion for the automatic selection of shape parameter Alpha) | |
 Public Member Functions inherited from smile::CDensity Public Member Functions inherited from smile::CDensity | |
| virtual double | Mass (const double r) const |
| returns mass inside given radius (approximately! not necessary to integrate density over sphere, just a rough estimate used e.g. in choosing radial nodes of Schwarzschild grid) | |
| virtual double | totalMass () const |
| returns estimated M(r=infinity) or -1 if mass is infinite | |
| double | getRadiusByMass (const double m) const |
| solves for Mass(r)=m | |
| void | getRadiiByMass (const vectord &masses, vectord *radii) const |
| solves for Mass(r)=m for an array of sorted values of m (more efficient than doing it one-by-one) | |
| bool | checkMassMonotonic () const |
| safety measure: check (roughly) that mass is increasing with radius | |
| bool | checkDensityNonzero () const |
| another safety measure: check that density doesn't drop to zero along any of three axes (important to assess spherical-harmonic approximation quality) | |
Static Public Member Functions | |
| static const char * | myName () |
Private Member Functions | |
| void | computeW (double lambda, double W[20]) const |
| compute the array of 20 coefficients W_{ijk} listed in Pfenniger(1984), in order of appearance in that paper; More... | |
Private Attributes | |
| const double | a |
| const double | b |
| const double | c |
| principal axis of ellipsoidal density | |
| const double | mass |
| const double | rho0 |
| total mass and central density of the model | |
| double | W0 [20] |
| pre-computed coefficients for lambda=0 | |
Additional Inherited Members | |
 Public Types inherited from smile::CDensity Public Types inherited from smile::CDensity | |
| enum | POTENTIALTYPE { PT_UNKNOWN, PT_DIRECT, PT_COMPOSITE, PT_COEFS, PT_NB, PT_BSE, PT_BSECOMPACT, PT_SPLINE, PT_CYLSPLINE, PT_LOG, PT_HARMONIC, PT_SCALEFREE, PT_SCALEFREESH, PT_SPHERICAL, PT_DEHNEN, PT_MIYAMOTONAGAI, PT_FERRERS, PT_PLUMMER, PT_ISOCHRONE, PT_PERFECTELLIPSOID, PT_NFW, PT_SERSIC, PT_EXPDISK, PT_ELLIPSOIDAL, PT_MGE } |
| list of all existing types of density or density/potential models, each of them implemented in its own class More... | |
| enum | SYMMETRYTYPE { ST_NONE = 0, ST_REFLECTION = 1, ST_PLANESYM = 2, ST_ZROTSYM = 4, ST_SPHSYM = 8, ST_TRIAXIAL = ST_REFLECTION | ST_PLANESYM, ST_AXISYMMETRIC = ST_TRIAXIAL | ST_ZROTSYM, ST_SPHERICAL = ST_AXISYMMETRIC | ST_SPHSYM, ST_DEFAULT = ST_TRIAXIAL } |
| Type of symmetry. More... | |
Ferrers density profile (with finite size R and density (1-(r/R)^2)^2).
The potential is calculated using expressions from Pfenniger(1984) with elliptic integrals, under assumption that p<q<1 strictly (will not work if any of two axes are equal).
|
inlinevirtual |
Return a pointer to a copy of this instance of potential.
A standard copy constructor or assignment is disabled because of different amount of data needed to be copied in different derived classes).
Implements smile::CPotential.
|
private |
compute the array of 20 coefficients W_{ijk} listed in Pfenniger(1984), in order of appearance in that paper;
| [in] | lambda | is zero inside the model and >0 outside; |
| [out] | W | is the array of 20 coefs |
|
virtual |
Compute forces and, optionally, force derivatives at a given point.
| [in] | xyz | - coordinates of the point to compute forces (array of 3 numbers) |
| [in] | t | - time to compute forces (matters only if potential is time-dependent) |
| [out] | force | - computed values of -d Phi/d x_i; output array must exist and contain N_DIM values. |
| [out] | forceDeriv | - if not NULL, then also compute the second derivatives of potential (which is a symmetric matrix of size N_DIM^2, thus the array must be of size N_DIM*(N_DIM+1)/2 ): first 3 values contain 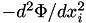 , second three contain mixed derivatives , second three contain mixed derivatives 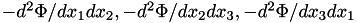 . . |
Implements smile::CPotential.
|
virtual |
Return potential at a given spatial point (possibly a time-varying one).
Implements smile::CPotential.
 1.8.8
1.8.8