 |
SMILE
v2.5
Schwarzschild Modelling Interactive expLoratory Environment
|
 |
SMILE
v2.5
Schwarzschild Modelling Interactive expLoratory Environment
|
common namespace for all core SMILE classes, functions and variables More...
Classes | |
| class | CAngMomOrbitFilteringFnc |
| Orbit filtering function that evaluates the z-component of angular momentum of an orbit. More... | |
| class | CBasicICGenerator |
| Base class for creating initial conditions for orbit library. More... | |
| class | CBasicInformation |
| parent class for data container objects used to store various data and exchange it between objects. More... | |
| class | CBasicIOSnapshot |
| The abstract class implementing reading and writing snapshots; derived classes take the filename as the argument of the constructor. More... | |
| class | CBasicOdeIntegrator |
| basic class for numerical integrators of ODE system More... | |
| class | CBasicOrbitFilteringFnc |
| Parent class for orbit filtering functions which evaluate whether an orbit should be used (in some calculation). More... | |
| class | CBasicOrbitRuntimeFnc |
| defines a routine which may be called after each integration timestep to perform user-specific data collection. More... | |
| class | CBasicOrbitRuntimeFncCreator |
| defines a basic class that creates some sort of runtime function for any orbit in the orbit library. More... | |
| class | CBasicQuadraticOptimizationSolver |
| The interface class for various third-party quadratic optimization solvers. More... | |
| class | CBasicSchwData |
| Base class for Schwarzschild Data objects. More... | |
| class | CBasicSchwModel |
| Base class for Schwarzschild Modelling driver objects. More... | |
| class | CBasicShellSchwData |
| Base class for storing kinematic information on a radial grid. More... | |
| class | CBasicSpectrumAnalyzer |
| Basic class for various methods of spectral analysis. More... | |
| class | CBinnedMassRefinementFnc |
| A mass refinement function is used to create an Nbody snapshot with unequal mass particles. More... | |
| class | CCentrophilicOrbitFilteringFnc |
| Orbit filtering function that selects centrophilic orbits. More... | |
| class | CChaosOrbitFilteringFnc |
| Orbit filtering function that evaluates chaotic properties of an orbit, based on threshold in frequency diffusion rate and in Lyapunov exponent value, multiplied by some predefined factor. More... | |
| struct | CConfigCore |
| Common configuration parameters for various tasks in CSmileCore. More... | |
| class | CConfigFile |
| Utility class for handling an INI file. More... | |
| struct | CConfigOrbit |
| general parameters for orbit integration and classification More... | |
| struct | CConfigPotential |
| structure that contains parameters for all possible potentials More... | |
| class | CDensity |
| Base class defining density model (without corresponding potential). More... | |
| class | CDensityEllipsoidal |
| Ellipsoidal model. More... | |
| class | CDensityExpDisk |
| Exponential disk with several variants of vertical profile and spiral structure. More... | |
| class | CDensityIsochrone |
| Isochrone density profile, triaxial. More... | |
| class | CDensityMGE |
| Multi-Gaussian expansion of a triaxial density profile. More... | |
| class | CDensityNFW |
| modified Navarro-Frenk-White density profile, triaxial. More... | |
| class | CDensityPerfectEllipsoid |
| Triaxial Perfect Ellipsoid density profile. More... | |
| class | CDensityPlummer |
| Plummer density profile, triaxial. More... | |
| class | CDensitySersic |
| Sersic profile, triaxial. More... | |
| class | CEnergyOrbitFilteringFnc |
| an example of evaluation function for multimass Nbody models, which sorts orbits in total energy More... | |
| class | CFileInputText |
| Class for reading data from a text file, possibly accompanied by binary files for the orbit library. More... | |
| class | CFileOutputText |
| Class for writing data to a text file, possibly accompanied by binary files for the orbit library. More... | |
| class | CICGeneratorDensityEllipsoidal |
| sample point coordinates using ellipsoidal equidensity surface approximation; this method is generally inferior to CICGeneratorDensitySampler More... | |
| class | CICGeneratorDensitySampler |
| sample points coordinates proportionally to local density, no velocities assigned More... | |
| class | CICGeneratorDensitySpherical |
| sample points from a spherical mass model More... | |
| class | CICGeneratorPrincipalPlane |
| Initial conditions from principal-plane start space. More... | |
| class | CICGeneratorRandomE |
| Initial conditions from random start space at given energy. More... | |
| class | CICGeneratorSchw |
| class that combines two samplers – for positions and velocities More... | |
| class | CICGeneratorStationary |
| Initial conditions from stationary start space. More... | |
| class | CICGeneratorVelocityEddington |
| sample velocities from the Eddington distribution function for equivalent spherical system More... | |
| class | CICGeneratorVelocityJeansAxi |
| sample velocities from Jeans equation for an axisymmetric system More... | |
| class | CICGeneratorVelocityJeansSph |
| sample velocities from the spherical Jeans equation More... | |
| class | CICGeneratorYalpha |
| Initial conditions from Y-alpha start space. More... | |
| class | CIOSnapshotGadget |
| GADGET snapshot format; needs UNSIO library. More... | |
| class | CIOSnapshotNemo |
| NEMO snapshot format. More... | |
| class | CIOSnapshotText |
| Text file with three coordinates, possibly three velocities and mass, space or tab-separated. More... | |
| class | CMassModel |
| class defining a spherical mass model. More... | |
| class | CMassModelAxisymmetric |
| Class for representing an axisymmetric density model. More... | |
| class | CMassModelAxisymmetricJeans |
| An extension of axisymmetric model which uses anisotropic Jeans equations to compute velocity dispersions in cylindrical coordinates. More... | |
| struct | CMatrix |
| interface for matrix storage. More... | |
| class | CMatrixBasicSchwModel |
| matrix interface class that passes the linear matrix elements from Schwarzschild model object to the solver More... | |
| struct | CMatrixDense |
| matrix interface class that actually holds the entire matrix and returns its elements More... | |
| struct | CMatrixDiagonal |
| matrix interface class that provides a diagonal matrix More... | |
| class | CNbodyExportStatWorker |
| Worker object that calculates statistics after Nbody export (currently, computes virial ratio). More... | |
| class | CNemoSnapshotWriter |
| helper class that writes NEMO-compatible snapshot file More... | |
| class | COdeIntegratorDOP853 |
| 8th order Runge-Kutta integrator from Hairer,Norsett&Wanner. More... | |
| class | COdeIntegratorHermite |
| Hermite integrator that uses information about force derivatives (jerk) in a 4th-order predictor-corrector scheme with only two force evaluations per timestep. More... | |
| class | COdeIntegratorIAS15 |
| 15-th order implicit Runge-Kutta scheme from Rein & Spiegel, 2014, MNRAS (adapted from Rebound). More... | |
| class | COdeIntegratorLeapfrog |
| low-order leapfrog method provided for treecode, which has discontinuous potential/forces and is poorly handled by Runge-Kutta integrator. More... | |
| class | COdeIntegratorOdeint |
| A template class for integrators based on the boost::numeric::odeint library. More... | |
| class | COdeSystem |
| basic class of ODE system used in the orbit integrator. More... | |
| class | COptimizationSolverBPMPD |
| Interface to the standalone program BPMPD. More... | |
| class | COptimizationSolverCVXOPT |
| Interface to the LP/QP solver CVXOPT written in Python. More... | |
| class | COptimizationSolverGLPK |
| Interface to the linear optimization solver GLPK. More... | |
| class | COrbit |
| Class for performing a single orbit integration. More... | |
| class | COrbitDesc |
| Container for orbit initial conditions and information objects. More... | |
| class | COrbitInformation |
| information object containing orbit analysis results. More... | |
| class | COrbitIntWorker |
| Worker object that runs single orbit integration in a separate thread. More... | |
| class | COrbitLauncher |
| An utility class that keeps all necessary data to start orbit integration. More... | |
| class | COrbitLibrary |
| The collection of orbits. More... | |
| class | COrbitLibWorker |
| Worker object that runs orbit library integration (for use in multiple parallel threads). More... | |
| class | COrbitRuntimePericenter |
| class for recording pericenter passages and analyzing their distribution to figure out if the orbit is centrophilic or not More... | |
| class | COrbitRuntimePericenterCreator |
| corresponding creator class for the pericenter analysis runtime function More... | |
| class | COrbitRuntimePoincare |
| compute the Poincare surface of section for an orbit. More... | |
| class | COrbitRuntimePoincareCreator |
| corresponding creator class for Poincare timestep fnc More... | |
| class | COrbitRuntimeSchwAngMomDist |
| Runtime function recording binned angular momentum distribution. More... | |
| class | COrbitRuntimeSchwAngMomDistCreator |
| corresponding creator class for angular momentum Schw runtime function More... | |
| class | COrbitRuntimeSchwDensGrid |
| Runtime function corresponding to classic 3d-grid Schwarzschild density data object. More... | |
| class | COrbitRuntimeSchwDensGridCreator |
| corresponding creator class for classic Schw runtime function More... | |
| class | COrbitRuntimeSchwDensSH |
| Runtime function corresponding to Schwatzschild density data object based on spherical-harmonic expansion of potential coefficients at radial grid points or for some radial basis functions (serves both variants) More... | |
| class | COrbitRuntimeSchwDensSHCreator |
| corresponding creator class for SH Schw runtime function More... | |
| class | COrbitRuntimeSchwKinemShell |
| Runtime function recording shell-based kinematic data (time spent in each shell and radial/tangential velocity dispersion in a shell). More... | |
| class | COrbitRuntimeSchwKinemShellCreator |
| corresponding creator class for shell-kinematic Schw runtime function More... | |
| class | COrbitRuntimeTrajectory |
| Class for recording orbit trajectory and doing various analysis (frequency, Lyapunov exponent, etc). More... | |
| class | COrbitRuntimeTrajectoryCreator |
| corresponding creator class for Trajectory analysis timestep fnc More... | |
| class | COrbitRuntimeTrajSample |
| Trajectory sampler (record a given number of unsorted points from the trajectory, to be used in creating an Nbody model from the Schwarzschild model). More... | |
| class | COrbitRuntimeTrajSampleCreator |
| corresponding creator class for trajectory sampler. More... | |
| class | COrbitRuntimeVelPerturber |
| class for applying random perturbations on velocity during orbit integration, mimicking the effect of two-body relaxation. More... | |
| class | COrbitRuntimeVelPerturberCreator |
| corresponding creator class for the velocity perturber runtime function More... | |
| class | COutputHookQProcess |
| Helper class to capture diagnostic and progress messages from running BPMPD process, and redirect them to the main SMILE program via my_message() More... | |
| class | CPericenterInformation |
| information object for pericenter data (distribution of angular momentum at pericenter passages) More... | |
| class | CPoincareInformation |
| information object to store points from the Poincare surface of section More... | |
| struct | CPointMassSet |
| An array of particles with positions, velocities and masses. More... | |
| class | CPointMassSetHandler |
| A helper object for computing the virial ratio for a set of point masses. More... | |
| struct | CPosPoint |
| Data-only class containing 3 coordinates. More... | |
| struct | CPosVelPoint |
| Data-only class containing 3 coordinates and 3 velocities. More... | |
| class | CPotential |
| base class for potential-density model including potential derivatives (=forces) More... | |
| class | CPotentialAxisymmetrized |
| A helper class for representing axisymmetrized versions of the given potential and/or density model. More... | |
| class | CPotentialBSE |
| basis-set expansion on the Zhao(1996) basis set (alpha models) More... | |
| class | CPotentialBSECompact |
| basis-set expansion using spherical Bessel functions (e.g.Allen et al.1990) More... | |
| class | CPotentialComposite |
| A composite potential which consists of several other potential types, plus a possible black hole and a rotating reference frame. More... | |
| class | CPotentialCylSpline |
| angular expansion of potential in azimuthal angle with coefficients being 2d spline functions of R,z More... | |
| class | CPotentialDehnen |
| Dehnen(1993) double power-law model. More... | |
| class | CPotentialDirect |
| Direct computation of potential for any density profile, using double integration over space. More... | |
| class | CPotentialFerrers |
| Ferrers density profile (with finite size R and density (1-(r/R)^2)^2). More... | |
| class | CPotentialHarmonic |
| Potential of a constant-density (harmonic) core with different frequencies of oscillations in three directions. More... | |
| class | CPotentialLog |
| Logarithmic potential, with possible core. More... | |
| class | CPotentialMiyamotoNagai |
| Miyamoto-Nagai axisymmetric potential; equivalent to Plummer if A=0. More... | |
| class | CPotentialNB |
| frozen N-body potential calculated by tree-code algorithm (based on hackcode1.c) More... | |
| class | CPotentialScaleFree |
| simple power-law potential/density pair: rho ~ r^-gamma More... | |
| class | CPotentialScaleFreeSH |
| angular expansion of scale-free potential in spherical harmonics More... | |
| class | CPotentialSH |
| parent class for all potential expansions based on spherical harmonics for angular variables More... | |
| class | CPotentialSpherical |
| Spherically-symmetric arbitrary density profile potential provided by CMassModel. More... | |
| class | CPotentialSpline |
| spherical-harmonic expansion of potential with coefficients being spline functions of radius More... | |
| class | CRadiusMassRefinementFnc |
| Another mass refinement function that aims to reduce the average mass of particles inside a given radius. More... | |
| class | CSchwarzschildWorker |
| Worker object that runs Schwarzschild model optimization solver. More... | |
| class | CSchwDataAngMomDist |
| Schwarzschild data object to manage kinematic data in the form of binned angular momentum distribution as a function of energy. More... | |
| class | CSchwDataBasicDens |
| Base class for density data objects. More... | |
| class | CSchwDataDensGrid |
| Base class for Schwarzschild data objects with grid-based density model. More... | |
| class | CSchwDataDensGridClassic |
| Schwarzschild data object for the classic grid-based density model. More... | |
| class | CSchwDataDensGridCylindrical |
| Schwarzschild data object for the grid-based density model in cylindrical geometry. More... | |
| class | CSchwDataDensSH |
| Base class for variants of Schwarzschild density data objects based on spherical-harmonic expansion of density of both target model and orbits. More... | |
| class | CSchwDataDensSHBSE |
| Variant of Schwarzschild density data object based on basis-set expansion in terms of Zhao(1995) basis set for density of both required profile and of orbits. More... | |
| class | CSchwDataDensSHMesh |
| Variant of Schwarzschild density data object based on spherical-harmonic expansion of potential coefficients at radial mesh points. More... | |
| class | CSchwDataKinemShell |
| Schwarzschild data object to handle (rudimentary) kinematic data, in the form of radial and tangential velocity dispersions in a number of radial shells. More... | |
| class | CSchwInformation |
| Information object that keeps density data for all variants of Schwarzschild data objects. More... | |
| class | CSchwModelQuadOpt |
| The class for performing Schwarzschild modelling of density and kinematic data from the theoretical point of view (without "observational errors"). More... | |
| class | CShellOrbitFilteringFnc |
| Orbit filtering function based on whether an orbit belongs to a given energy shell. More... | |
| class | CSmileConsole |
| The console version of application is implemented in this class. More... | |
| class | CSmileCore |
| Class that provides all core computational functionality. More... | |
| class | CSmileGUI |
| The main GUI window class. More... | |
| struct | CSpectralLine |
| Definition of one spectral line in one-dimensional time series (nothing to do with spectroscopy!:) More... | |
| class | CSpectrumAnalyzerHunter |
| Spectrum analysis based on Hunter's DFT method. More... | |
| class | CSplineApprox |
| Penalized linear least-square fitting problem. More... | |
| class | CTrajSampleInformation |
| information object keeping a trajectory sample (to generate an N-body model from a Schwarzschild model) More... | |
| struct | CTriangle |
| defines a triangle with indexed vertices referring to points from the trajectory More... | |
| class | CTriangThread |
| Helper thread that performs Delaunay triangulation with the help of external program 'qdelaunay'. More... | |
| class | CVelPerturberInformation |
| information object for velocity perturber class (keep track of accumulated energy change due to relaxation) More... | |
| class | QwtPlotPicker2 |
| helper class for displaying coordinates on a 2d plot picker with scientific numeric format string More... | |
| class | QwtPlotZoomer2 |
| helper class for displaying coordinates on a 2d plot zoomer with scientific numeric format string More... | |
| class | StringVariant |
| The StringVariant class is a simple string-based variant implementation that allows the user to easily convert between simple numeric/string types. More... | |
Typedefs | |
|
typedef std::pair< float, CPointMassSet< float > ::ElemType > | CPointMassEnergy |
|
typedef boost::numeric::odeint::bulirsch_stoer_dense_out < OdeStateType > | StepperBS |
| Implementation of Bulirsch-Stoer method with dense output. | |
|
typedef boost::numeric::odeint::dense_output_runge_kutta < boost::numeric::odeint::controlled_runge_kutta < boost::numeric::odeint::runge_kutta_dopri5 < OdeStateType > , my_error_checker > > | StepperDP5 |
| Implementation of the Dormand-Prince 5th order Runge-Kutta method with adaptive timestep and dense output. | |
|
typedef boost::numeric::odeint::controlled_runge_kutta < boost::numeric::odeint::runge_kutta_cash_karp54 < OdeStateType > , my_error_checker > | StepperCK5 |
| Implementation of the Cash-Karp 5th order Runge-Kutta method with adaptive timestep. | |
|
typedef boost::numeric::odeint::controlled_runge_kutta < boost::numeric::odeint::runge_kutta_bogacki_shampine32 < OdeStateType > , my_error_checker > | StepperBS3 |
| Implementation of the Bogacki-Shampine 3rd order Runge-Kutta method with adaptive timestep. | |
|
typedef boost::numeric::odeint::runge_kutta4 < OdeStateType > | StepperRK4 |
| Implementation of the standard fixed-timestep 4th order Runge-Kutta method. | |
|
typedef boost::numeric::odeint::symplectic_rkn_sb3a_mclachlan < OdeStateType > | StepperSympl4 |
| Implementation of the fixed-timestep 4th order symplectic Runge-Kutta method. | |
|
typedef std::vector < CSpectralLine > | CSpectrum |
| An array of spectral lines in one-dimensional time series. | |
| typedef std::complex< double > | complexd |
| convenience shorthand for a complex number | |
|
typedef std::map < CDensity::POTENTIALTYPE, const char * > | PotentialNameMapType |
| lists all 'true' potentials, i.e. those providing a complete density-potential(-force) pair | |
|
typedef std::map < CDensity::POTENTIALTYPE, const char * > | DensityNameMapType |
| lists all analytic density profiles (including those that don't have corresponding potential, but excluding general-purpose expansions) | |
|
typedef std::map < CDensity::SYMMETRYTYPE, const char * > | SymmetryNameMapType |
| lists available symmetry types | |
| typedef void | show_message_type (const std::string &origin, const std::string &message) |
| a function that shows a given message string. More... | |
------- define several convenience types ------- | |
| typedef float | sdNumType |
| how to store orbit data for Schwarzschild modelling (float or double) | |
| typedef float | smNumType |
| how to handle Schwarzschild modelling data internally (float or double) | |
| typedef std::vector< double > | vectord |
| vector of double values | |
| typedef std::vector< float > | vectorf |
| vector of float values | |
| typedef std::pair< double, double > | paird |
| a pair of doubles | |
| typedef std::vector< paird > | vectorpd |
| vector of pairs of doubles | |
| typedef std::pair< float, float > | pairf |
| a pair of floats | |
| typedef std::vector< pairf > | vectorpf |
| vector of pairs of floats | |
| typedef vectord | OdeStateType |
| container for the array of variables used in ODE integration | |
------- Definition of base (abstract) classes -------- | |
|
typedef std::vector< const CBasicSchwData * > | vectorSchwData |
| array of heterogeneous data objects to be used simultaneously in the modelling | |
Functions | |
| void | readConfigPotential (const CConfigFile &configReader, CConfigPotential *ptrConfigPotential, unsigned int potGroup=0) |
| Read the configuration data from the INI file which is contained in one [Potential*] section. More... | |
| void | writeConfigPotential (CConfigFile &configWriter, const CConfigPotential *ptrConfigPotential, unsigned int potGroup=0) |
| Write the configuration data for one potential component into the INI file. More... | |
| void | readConfigOrbit (const CConfigFile &configReader, CConfigOrbit *ptrConfigOrbit) |
| Read configuration data for orbit integration (a subset of parameters contained in the [Orbit] section of INI file). More... | |
| void | writeConfigOrbit (CConfigFile &configWriter, const CConfigOrbit *ptrConfigOrbit) |
| Write configuration data for orbit integration to INI file. More... | |
| QString | getDirName (const QString &fileName) |
| convenience function, return file path ending with "/" | |
| void | my_message_gui (const std::string &origin, const std::string &message) |
| display the message string in the bottom-right text area of the main window | |
| void | my_error_gui (const std::string &origin, const std::string &message) |
| pop up a message box with the error string | |
| CShellOrbitFilteringFnc | shellFilter (const vectorSchwData &schwData, const CPotential *potential, int numShell, double *shellEnergy=NULL) |
| double | densSampler (double x[N_DIM], size_t, void *param) |
| double | computeEnergyCutoff (const CPotential *potential, const CPointMassSet< float > &points) |
| std::vector< std::string > | initFormatsIOSnapshot () |
| create a list of all IO snapshot formats available at compile time | |
| CBasicIOSnapshot * | createIOSnapshotRead (const std::string &fileName) |
| creates an instance of appropriate snapshot reader, according to the file format determined by reading first few bytes, or NULL if a file doesn't exist | |
| CBasicIOSnapshot * | createIOSnapshotWrite (const std::string &fileFormat, const std::string &fileName, const std::string &header="") |
| creates an instance of snapshot writer for a given format name, or NULL if the format name string is incorrect | |
| const CDensity * | createDensity (const CConfigPotential *config) |
| create a density model according to the parameters. More... | |
| const CPotential * | createPotential (CConfigPotential *config) |
| create an instance of CPotential according to the parameters passed. More... | |
| template<typename NumT > | |
| const CPotential * | createPotentialFromPoints (const CConfigPotential *configPotential, const CPointMassSet< NumT > *points) |
| create a potential of a generic expansion kind from a set of point masses. More... | |
| template const CPotential * | createPotentialFromPoints (const CConfigPotential *configPotential, const CPointMassSet< float > *points) |
| template const CPotential * | createPotentialFromPoints (const CConfigPotential *configPotential, const CPointMassSet< double > *points) |
| const CPotential * | readPotential (CConfigPotential *configPotential) |
| load a potential from a text or snapshot file. More... | |
| bool | writePotential (const std::string &fileName, const CPotential *potential) |
| write potential expansion coefficients to a text file. More... | |
| CMassModel * | createMassModel (const CDensity *density, int numNodes=NUM_RADIAL_POINTS_SPHERICAL_MODEL, double Rmin=0, double Rmax=0, const CMassModel *poten=NULL) |
| create an equivalent spherical mass model for the given density profile. More... | |
| double | int_generic (double x, void *params) |
| double | int_generic_f (double x, void *params) |
| double | int_densproj (double z, void *params) |
| double | int_losvd (double z, void *params) |
| double | find_generic (double x, void *params) |
| double | find_alpha (double a, void *params) |
| double | find_rcirc (double r, void *params) |
| double | find_vrad (double r, void *params) |
| double | find_gamma_out (double y, void *params) |
| double | int_mass (double r, void *params) |
| double | int_generic_dif (double r, void *params) |
| double | int_generic_dif1 (double s, void *params) |
| double | int_Torb (double s, void *params) |
| double | find_diffusion_limit (double scaledE, void *param) |
| template<typename NumT > | |
| bool | sortfnc (const std::pair< NumT, NumT > &elem1, const std::pair< NumT, NumT > &elem2) |
| double | getfitquality (double totalmass, void *params) |
| template<typename NumT > | |
| bool | fitMassModel (const std::vector< NumT > &particlecoord,const std::vector< NumT > &particlevalue,std::vector< double > *gridcoord,std::vector< double > *gridvalue,double smoothing=0,size_t gridsize=0,size_t nskip=0,CMassModel::FITTYPE ftype=CMassModel::FT_MASS,const CMassModel *model=NULL) |
| Function that computes the input data for a spherical mass model from a given array of particles. More... | |
| template bool | fitMassModel (const std::vector< double > &particlecoord, const std::vector< double > &particlemass, std::vector< double > *gridcoord, std::vector< double > *gridmass, double smoothing, size_t gridsize, size_t nskip, CMassModel::FITTYPE ftype, const CMassModel *model) |
| template bool | fitMassModel (const std::vector< float > &particlecoord, const std::vector< float > &particlemass, std::vector< double > *gridcoord, std::vector< double > *gridmass, double smoothing, size_t gridsize, size_t nskip, CMassModel::FITTYPE ftype, const CMassModel *model) |
| int | glpk_output_hook (void *tmp, const char *data) |
| helper routine to capture debug and progress messages from GLPK and redirect them to my_message() | |
| void | glpk_error_harmless (void *) |
| change the default behaviour on GLPK error to something that doesn't crash program right away | |
| static PyObject * | redirection_stdoutredirect (PyObject *, PyObject *args) |
| helper routine to record messages from Python interpreter and pass them to my_message() | |
| template<typename NumT > | |
| PyObject * | initPyMatrix (const CMatrix< NumT > *srcmatrix) |
| helper routine to create CVXOPT-compatible dense or sparse matrix | |
| void | tuneNonPrime (size_t *n) |
| utility function to ensure that n does not have large prime divisors. More... | |
| bool | computeNperiods (const CConfigOrbit *configOrbit, const CPotential *potential, const CPosVelPoint< double > &initCond, int coord_section, int Nperiods, CPosVelPoint< double > *endCond) |
| starting from the given initial conditions in a plane x[coord_section]=0, x'[coord_section]!=0, integrate an orbit until it crosses the same plane for N-th time. More... | |
| int | gsl_wrapper_computeNperiods (const gsl_vector *initC, void *param, gsl_vector *values, gsl_matrix *jac) |
| int | gsl_wrapper_computeNperiods_f (const gsl_vector *initC, void *param, gsl_vector *values) |
| int | gsl_wrapper_computeNperiods_df (const gsl_vector *initC, void *param, gsl_matrix *jac) |
| bool | findPeriodicOrbit (const CPotential *potential, double Omega, unsigned int coord_section, unsigned int coord_first, bool threeD, unsigned int Nperiods, CPosVelPoint< double > *IC) |
| Utility function to find a periodic orbit in the plane coord1:coord2 which is closest to the provided initial conditions. More... | |
| template<typename NumT > | |
| double | computeTimeUnit (const CPotential *potential, const CPosVelPoint< NumT > &initCond) |
| Utility function to get the period of long-axis orbit in the given potential with the energy E corresponding to initial conditions. More... | |
| template double | computeTimeUnit (const CPotential *potential, const CPosVelPoint< float > &initCond) |
| template double | computeTimeUnit (const CPotential *potential, const CPosVelPoint< double > &initCond) |
| double | findLcirc2 (const CPotential *potential, const CPosVelPoint< double > &initCond) |
| Utility function to find approximate L^2 of a circular orbit with given energy. | |
| double | findLambda (double lambda, void *param) |
| gsl_spline * | init_spline_hypergeom (double m) |
| double | my_hypergeom (double x, const gsl_spline *spl) |
| double | integrandPotentialDirectDensity (double phi, void *param) |
| double | LegendreQ (double m, double x, const gsl_spline *spl) |
| Legendre function Q(m,x) expressed through Gauss hypergeometric function. | |
| double | IntBessel (double m, double a, double b, double c, const gsl_spline *spl, double eps2=0) |
| $^ J_m(a*x)*J_m(b*x)*exp(-|c|*x) dx / (()*(m+1/2)/(m+1))$ | |
| double | integrandPotentialDirectVar2 (double var, void *params) |
| double | integrandPotentialDirectVar1 (double var, void *params) |
| double | integrandPotAvgPhi (double phi, void *param) |
| void | interp2d_spline_eval_all (const interp2d_spline *interp, const double x, const double y, double *z, double *z_x, double *z_y, double *z_xx, double *z_xy, double *z_yy) |
| optimized routine for computing 2d spline value, first and second derivatives at once (to save time) | |
| void | initPotentialAndSymmetryNameMap () |
| create a correspondence between names and enum identifiers for potential, density and symmetry types | |
| std::string | undecorateFunction (std::string origin) |
| remove common prefix if it is the namespace "smile::", and remove function signature from GCC PRETTY_FUNCTION | |
| void | my_stderr_show_message (const std::string &origin, const std::string &message) |
| default routine that dumps text messages to stderr More... | |
| void | gsl_verbose_error_handler (const char *reason, const char *file, int line, int gsl_errno) |
| error reporting routine which is invoked from inside GSL. More... | |
| std::string | pp (double num, unsigned int width) |
| Pretty-print: convert floating-point or integer numbers to a string of fixed length. | |
| void | splitString (const std::string &src, const std::string &delim, std::vector< std::string > *result) |
| routine that splits one line from a text file into several items. More... | |
| bool | ends_with_str (const std::string &str, const std::string &end) |
| routine that checks if a string ends with another string | |
| bool | strings_equal (const std::string &str1, const std::string &str2) |
| routine that compares two strings in a case-insensitive way | |
| bool | strings_equal (const std::string &str1, const char *str2) |
| overloaded routine that compares two strings in a case-insensitive way | |
| void | randomize () |
| initialize random number generator using current time | |
| double | getRandomNumber () |
| get a random number uniformly distributed in [0..1] | |
| void | getNormalRandomNumbers (double *num1, double *num2) |
| generates two random numbers with normal distribution (zero mean, dispersion=1) | |
| void | getRandomUnitVector (double *x, double *y, double *z) |
| generates 2d or 3d random vector with unit length; if z==NULL then create 2d vector, else 3d | |
| double | findRoot (gsl_function *F, double xlower, double xupper, double xdefault=0, double reltoler=1e-8) |
| wrapper for GSL root solver running multiple iterations until the convergence is achieved. More... | |
| double | findMin (gsl_function *F, double xlower, double xupper, double xinit, double reltoler=1e-8) |
| wrapper for GSL minimization routine to find local minimum enclosed between xlower < xinit < xupper. More... | |
| double | findMinGuess (gsl_function *F, double xlower, double xupper, double reltoler=1e-8) |
| wrapper for GSL minimization routine to find local minimum enclosed between xlower < xupper. More... | |
| void | createNonuniformGrid (size_t nnodes, double xmin, double xmax, bool zeroelem, std::vector< double > *grid) |
| generates a grid with exponentially growing spacing. More... | |
| static void | makegrid (std::vector< double >::iterator begin, std::vector< double >::iterator end, double startval, double endval) |
| creation of a grid with minimum guaranteed number of input points per bin | |
| void | createAlmostUniformGrid (const std::vector< double > &srcpoints, size_t minbin, size_t *gridsize, std::vector< double > *grid) |
| creates an almost uniform grid so that each bin contains at least minbin points from input array. More... | |
| double | gsl_spline_evalx (const gsl_spline *spline, double x, gsl_interp_accel *a) |
| modified functions for spline evaluation that extrapolate last segment linearly beyond the definition region | |
| double | gsl_spline_eval_derivx (const gsl_spline *spline, double x, gsl_interp_accel *a) |
| double | gsl_spline_eval_deriv2x (const gsl_spline *spline, double x, gsl_interp_accel *a) |
| void | cspline_eval_all (const gsl_spline *spline, double x, double *val, double *der, double *der2) |
| convenience function that evaluates cubic spline value, derivative and 2nd derivative at once (faster than doing it separately) | |
| bool | check_spline_monotonic (const gsl_spline *spline) |
| check if a cubic spline is everywhere monotonic | |
| gsl_spline * | copyspline (const gsl_spline *src) |
| create a copy of a spline by allocating new memory to all internal arrays and copying their data | |
| double | getVal (const gsl_matrix *deriv, size_t row, size_t col) |
| convenience function returning values from band matrix or zero if indexes are outside the band | |
| static void | my_error (const std::string &origin, const std::string &message) |
| the interface routine for error reporting (redirects the call to my_error_ptr if it is defined) | |
| static void | my_message (const std::string &origin, const std::string &message) |
| the interface routine for message reporting (redirects the call to my_message_ptr if it is defined) | |
| template<typename NumberType , typename ValueType > | |
| NumberType | convertTo (ValueType val) |
| a shorthand function to convert a string to a number | |
| template<typename ValueType > | |
| bool | convertToBool (ValueType val) |
| a shorthand function to convert a string to a bool | |
| template<typename ValueType > | |
| std::string | convertToString (ValueType val) |
| a shorthand function to convert a number to a string | |
| static const char * | convertToString (bool val) |
| a shorthand to convert a bool to a string | |
| template<typename ValueType > | |
| std::string | convertToString (ValueType val, unsigned int width) |
| a shorthand function to convert a number to a string with a given precision | |
--- non-class utility routines --- | |
| const char * | getICgenNameByType (CBasicICGenerator::ICTYPE ICType) |
| return the name for a given IC generator data type | |
| CBasicICGenerator::ICTYPE | getICgenTypeByName (const std::string &ICName) |
| return IC generator data type associated with the text name | |
| const char * | getIntegratorNameByType (CBasicOdeIntegrator::STEPPERKIND integratorType) |
| CBasicOdeIntegrator::STEPPERKIND | getIntegratorTypeByName (const std::string &integratorName) |
| return integrator type associated with the text name | |
Various convenience functions for potential | |
| double | longAxisRadius (const CPotential *potential, double E, double Omega=0) |
| Convenience function to find the maximal elongation of long-axis orbit of given energy. More... | |
| double | findIntersection (const CPotential *potential, double E, double X, double Y, double Z, double Omega=0) |
| Find the location of the equipotential surface in a given direction: solve for k such that Phi(k*X, k*Y, k*Z) = E . More... | |
| void | findLagrangianPoints (const CPotential *potential, double Omega, double *L1X, double *L4Y) |
| Find the location of maximum of the effective potential in rotating frame; output the distance to such point along x axis in L1X and along y axis in L4Y (Lagrangian points) | |
| template<typename NumT > | |
| double | totalEnergy (const CPotential *potential, const CPosVelPoint< NumT > &point, double Omega=0, double t=0) |
| Convenience function to return the total energy (or Jacobi integral if Omega!=0) for a given position+velocity point. More... | |
------- Miscellaneous functions -------- | |
| const char * | getSchwDataNameByType (CBasicSchwData::SCHWDATATYPE dataType) |
| Correspondence between Schw.model names and types. More... | |
| CBasicSchwData::SCHWDATATYPE | getSchwDataTypeByName (const std::string &DataName) |
| return Schwarzschild data type associated with the text name (Classic by default) | |
| CBasicOrbitRuntimeFncCreator * | createSchwCreator (const CBasicSchwData *schwData) |
| convenience function to return an orbit runtime function creator associated with a given Schwarzschild data class | |
| void | createRadialGrid (const CDensity *density, size_t numShells, double totalMass, double innerShellMass, double outerShellMass, vectord *grid) |
| convenience function that creates a "non-uniform grid" in mass and returns array of corresponding radii. | |
Variables | |
| const CConfigCore | defaultConfigCore |
| default configuration parameters | |
| volatile int | nthreads =0 |
| QMutex | mutexL |
| QMutex | mutexC |
| CSmileGUI * | GUI |
| The global variable for the instance of GUI object is used to redirect text messages to it. | |
| std::vector< std::string > | formatsIOSnapshot = initFormatsIOSnapshot() |
| list of all available IO snapshot formats, initialized at module start according to the file format supported at compile time | |
| const size_t | splJ_sizex =31 |
| const size_t | splJ_sizey =100 |
| const double | splJ_facty =5.0 |
| static jmp_buf | err_buf |
| static bool | memoryError =false |
| flag set if Python reports about an out-of-memory error | |
| static PyMethodDef | RedirectionMethods [] |
| const CConfigOrbit | defaultConfigOrbit |
| default parameters for orbit integration and classification More... | |
| PotentialNameMapType | PotentialNames |
| DensityNameMapType | DensityNames |
| SymmetryNameMapType | SymmetryNames |
| bool | mapinitialized = false |
| show_message_type * | my_error_ptr = &my_stderr_show_message |
| global variable to the routine that displays errors (if it is NULL then nothing is done) | |
| show_message_type * | my_message_ptr = &my_stderr_show_message |
| global variable to the routine that shows information messages (emitted by various time-consuming routines to display progress), if NULL then no messaging is done More... | |
| gsl_rng * | randgen =gsl_rng_alloc(gsl_rng_default) |
| an instance of global random number generator | |
| const gsl_interp_type * | gsl_interp_ccspline |
| a variant of cubic spline with given values of first derivative at endpoints. More... | |
----------- Parameters for potential and force evaluation ------------- | |
| const unsigned int | N_DIM =3 |
| Dimensions of the real world. DO NOT CHANGE (unless you are God)! | |
| const size_t | MAX_NCOEFS_RADIAL =101 |
| max number of basis-function expansion members (radial and angular). | |
| const size_t | MAX_NCOEFS_ANGULAR =51 |
| const double | EPSREL_DENSITY_INT =1e-4 |
| relative accuracy of density computation | |
| const double | EPSREL_POTENTIAL_INT =1e-6 |
| relative accuracy of potential computation (integration tolerance parameter) | |
| const double | EPSABS_POTENTIAL_INT =1e-15 |
| absolute error in potential computation (supercedes the relative error in case of very small coefficients) | |
| const double | EPSREL_MASS_INT =1e-2 |
| relative accuracy of cell mass computation in Schwarzschild modelling | |
| const double | MIN_RADIUS_CUTOFF =1e-6 |
| inner and outer cutoff radii appearing in calculations of M(r), r(E) and related | |
| const double | MAX_RADIUS_CUTOFF =1e+6 |
| const double | SPLINE_MIN_RADIUS =1e-10 |
| auxiliary softening parameter for Spline potential to avoid singularities in some odd cases | |
------------- Orbit integration parameters ---------- | |
| const size_t | NUMSTEP_MAX =static_cast<size_t>(1e7) |
| maximal number of integration steps (in case something went wrong with the timestep, do not dead-lock) | |
| const double | TREECODE_TIMESTEP_BH_FACTOR =0.2 |
| for Nbody treecode, another safety measure is to decrease timestep near BH by this factor (in the leap-frog integrator) | |
| const size_t | TREECODE_DENSITY_KTH_NEIGHBOUR =32 |
| density estimate in treecode by using distance to k-th neighbour; if using spline kernel, put more (>~20), since this kernel is more concentrated | |
| const double | LYAP_DEV_INIT =1e-10 |
| initial value of orbit separation (if variational equation is not used and two nearby trajectories are followed) | |
| const double | LYAP_DEV_MAX =1e-6 |
| if distance between nearby trajectories reaches the following threshold, renormalize it back to LYAP_DEV_INIT | |
| const double | LYAP_VAREQ_DEV_MAX =1e5 |
| if variational equation is used, perform renormalization of deviation vector if greater than threshold (only for numerical convenience, since the var.equation is anyway linear) | |
------------- Orbit spectral analysis and classification options ---------- | |
| const size_t | MAX_FFT_PRIME =41 |
| orbit spectrum is calculated by fast fourier transform from GSL. More... | |
| const double | FREQ_PRECISE_THRESHOLD =5.0 |
| Use Hunter's method for precise determination of frequency of a given line if the spectrum neat its maximum is reasonably similar to that of a single line. More... | |
| const size_t | MAX_SPECTRAL_LINES =10 |
| maximal number of spectral lines in each coordinate to search for | |
| const double | MIN_SPECTRAL_LINE_AMPLITUDE =1e-2 |
| do not use lines which amplitude is X times lower than amplitude of the leading line | |
| const double | FREQ_ACCURACY =0.1 |
| accuracy of frequency comparison (to determine resonances), in units of Nyquist frequency | |
| const double | FREQ_DIFF_REG_MIN =1e-6 |
| minimum value of freq.diff threshold separating regular and chaotic orbits (actual threshold depends on integration interval) | |
| const int | FREQ_RES_2 =10 |
| maximum order of m:n frequency resonance (or linear dependence of three frequencies) | |
| const int | FREQ_RES_3 =10 |
| maximum order of l*a+m*b+n*c frequency commensurability (thin orbit) | |
| const double | OC_PYRAMID_AVGZ =0.1 |
| threshold value of |<z>| / rmax to classify orbit as pyramid or saucer | |
| const size_t | MAX_POINCARE_POINTS =10000 |
| max number of points in Poincare section | |
| const double | PERICENTER_FIT_FRACTION =0.1 |
| fraction of closest pericenter passages which are used in fitting | |
| const int | PERICENTER_FIT_MIN_POINTS =10 |
| minimum number of points used in pericenter fitting | |
| const int | PERICENTER_FIT_MAX_POINTS =50 |
| defines max points in pericenter fitting | |
------------- Orbit library / Schwarzschild modelling options --------------- | |
sample initial conditions out to a radius containing this fraction of mass | |
| const double | MAX_LAGRANGIAN_RADIUS =0.999 |
| const size_t | NUM_RADIAL_POINTS_SPHERICAL_MODEL =100 |
| number of radial points used in createMassModel | |
| const double | SCHW_MAX_CELLMASS_REL_DIFFERENCE =1e-2 |
| max relative difference in cell mass to consider the cell as feasible: abs(1-Mcell/Mcellrequired) | |
| const double | SCHW_MIN_ORBIT_WEIGHT_USED =1e-4 |
| min orbit weight to use the orbit after the optimization problem solved (divide by number of orbits!) | |
| const double | NBEXPORT_XV_FACTOR =10.0 |
| Nbody export: relative importance of nullifying total angular momentum at the expense of pericenter velocity * pericenter offset. | |
-------------- Options for GUI and high-level operations ------------ | |
| const size_t | NUM_POINTS_PLOT_EQUIPOTENTIAL =100 |
| number of points in curve representing equipotential surface (actually *4) | |
| const size_t | NUM_POINTS_PLOT_POINCARE =100 |
| number of points in outer bounding curve in Poincare section (actually *4) | |
| const double | SCHW_MIN_ORBIT_WEIGHT_DISPLAYED =1e-4 |
| min. weight of orbit to be displayed in GUI | |
| const double | PLOT_FREQ_MAX_F =5.0 |
| width of frequency spectrum displayed (in units of orbital frequency) | |
-------- Various convenience definitions -------- | |
|
typedef std::vector < CBasicOrbitRuntimeFnc * > | vectorRuntimeFncs |
| convenience definition of vectors of timestep functions and function creators | |
|
typedef std::vector< const CBasicOrbitRuntimeFncCreator * > | vectorRuntimeFncCreators |
|
typedef std::vector< const CBasicInformation * > | vectorInformation |
|
typedef std::vector < CPosVelPoint< double > > | CPosVelDataDouble |
| convenience definitions of templated vectors containing particle sets with/without masses | |
|
typedef std::vector < CPosVelPoint< float > > | CPosVelDataFloat |
| double | pow_2 (double x) |
| convenience shorthand for squaring a number | |
| float | pow_2 (float x) |
| size_t | pow_2 (size_t x) |
| template<typename T1 , typename T2 > | |
| bool | comparepair (const std::pair< T1, T2 > &val1, const std::pair< T1, T2 > &val2) |
| convenience function for sorting arrays of std::pair<float/double, something> | |
Correspondence between potential/density names and corresponding classes | |
| const CConfigPotential | defaultConfigPotential |
| default parameters for all possible potentials More... | |
| const char * | getPotentialNameByType (CDensity::POTENTIALTYPE type) |
| return the name of the potential of a given type, or empty string if unavailable | |
| const char * | getDensityNameByType (CDensity::POTENTIALTYPE type) |
| return the name of the density of a given type, or empty string if unavailable | |
| const char * | getSymmetryNameByType (CDensity::SYMMETRYTYPE type) |
| return the name of the symmetry of a given type, or empty string if unavailable | |
| CDensity::POTENTIALTYPE | getPotentialTypeByName (const std::string &PotentialName) |
| return the type of the potential model by its name, or PT_UNKNOWN if unavailable | |
| CDensity::POTENTIALTYPE | getDensityTypeByName (const std::string &DensityName) |
| return the type of the density model by its name, or PT_UNKNOWN if unavailable | |
| CDensity::SYMMETRYTYPE | getSymmetryTypeByName (const std::string &SymmetryName) |
| return the type of symmetry by its name, or ST_DEFAULT if unavailable | |
| const char * | coefFileExtension (CDensity::POTENTIALTYPE pottype) |
| return file extension for writing the coefficients of potential of the given type | |
| CDensity::POTENTIALTYPE | coefFileType (const std::string &fileName) |
| find potential type by file extension | |
common namespace for all core SMILE classes, functions and variables
| typedef void smile::show_message_type(const std::string &origin, const std::string &message) |
a function that shows a given message string.
implementation is program-specific, may dump text to stderr or do something else.
| [in] | origin | is the name of calling function |
| [in] | message | is the text to be displayed |
| bool smile::computeNperiods | ( | const CConfigOrbit * | configOrbit, |
| const CPotential * | potential, | ||
| const CPosVelPoint< double > & | initCond, | ||
| int | coord_section, | ||
| int | Nperiods, | ||
| CPosVelPoint< double > * | endCond | ||
| ) |
starting from the given initial conditions in a plane x[coord_section]=0, x'[coord_section]!=0, integrate an orbit until it crosses the same plane for N-th time.
Return the final coordinates at this crossing.
| double smile::computeTimeUnit | ( | const CPotential * | potential, |
| const CPosVelPoint< NumT > & | initCond | ||
| ) |
Utility function to get the period of long-axis orbit in the given potential with the energy E corresponding to initial conditions.
It is given by the integral 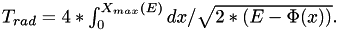
| void smile::createAlmostUniformGrid | ( | const std::vector< double > & | srcpoints, |
| size_t | minbin, | ||
| size_t * | gridsize, | ||
| std::vector< double > * | grid | ||
| ) |
creates an almost uniform grid so that each bin contains at least minbin points from input array.
input points are in srcpoints array and MUST BE SORTED in ascending order (assumed but not cheched). grid must point to an existing (but possibly uninitialized) vector which will have length at most gridsize. NB: in the present implementation, the algorithm is not very robust and works well only for gridsize*minbin << srcpoints.size, assuming that 'problematic' bins only are found close to endpoints but not in the middle of the grid.
| const CDensity * smile::createDensity | ( | const CConfigPotential * | config | ) |
create a density model according to the parameters.
This only deals witj finite-mass models, including some of the CPotential descendants.
| [in] | configPotential | contains the parameters (density type, mass, shape, etc.) |
| CMassModel * smile::createMassModel | ( | const CDensity * | density, |
| int | numNodes = NUM_RADIAL_POINTS_SPHERICAL_MODEL, |
||
| double | Rmin = 0, |
||
| double | Rmax = 0, |
||
| const CMassModel * | poten = NULL |
||
| ) |
create an equivalent spherical mass model for the given density profile.
| [in] | density | is the non-spherical density model to be approximated |
| [in] | poten | (optional) another spherical mass model that provides the potential (if it is not given self-consistently by this density profile). |
| [in] | numNodes | specifies the number of radial grid points in the M(r) profile |
| [in] | Rmin | is the radius of the innermost non-zero grid node (0 means auto-select) |
| [in] | Rmax | is the radius outermost grid node (0 means auto-select) |
| void smile::createNonuniformGrid | ( | size_t | nnodes, |
| double | xmin, | ||
| double | xmax, | ||
| bool | zeroelem, | ||
| std::vector< double > * | grid | ||
| ) |
generates a grid with exponentially growing spacing.
x[k] = (exp(Z k) - 1)/(exp(Z) - 1), and the value of Z is computed so the the 1st element is at xmin and last at xmax.
| [in] | nnodes | – total number of grid points |
| [in] | xmin,xmax | – location of the first and the last node |
| [in] | zeroelem | – if true, 0th node is at zero (otherwise at xmin) |
| [out] | grid | – pointer to an existing array which will be overwritten by this routine |
| const CPotential * smile::createPotential | ( | CConfigPotential * | config | ) |
create an instance of CPotential according to the parameters passed.
| [in,out] | config | specifies the potential parameters, which could be modified, e.g. if the potential coefficients are loaded from a file. Massive black hole (config->Mbh) is not included in the potential (the returned potential is always non-composite) |
| const CPotential * smile::createPotentialFromPoints | ( | const CConfigPotential * | configPotential, |
| const CPointMassSet< NumT > * | points | ||
| ) |
create a potential of a generic expansion kind from a set of point masses.
| [in] | configPotential | contains the parameters (potential type, number of terms in expansion, etc.) |
| [in] | points | is the array of particles that are used in computing the coefficients; if potential type is PT_NB, then an instance of tree-code potential is created. |
| double smile::findIntersection | ( | const CPotential * | potential, |
| double | E, | ||
| double | X, | ||
| double | Y, | ||
| double | Z, | ||
| double | Omega = 0 |
||
| ) |
Find the location of the equipotential surface in a given direction: solve for k such that Phi(k*X, k*Y, k*Z) = E .
| double smile::findMin | ( | gsl_function * | F, |
| double | xlower, | ||
| double | xupper, | ||
| double | xinit, | ||
| double | reltoler = 1e-8 |
||
| ) |
wrapper for GSL minimization routine to find local minimum enclosed between xlower < xinit < xupper.
xinit is the initial guess and the value to be returned in the case of error.
| double smile::findMinGuess | ( | gsl_function * | F, |
| double | xlower, | ||
| double | xupper, | ||
| double | reltoler = 1e-8 |
||
| ) |
wrapper for GSL minimization routine to find local minimum enclosed between xlower < xupper.
employed when the initial minimum location is not really known but suspected to exist in the interval. performs binary search to find the initial minimum location and then calls GSL routines to refine it. if the interval doesn't enclose minimum then one of the endpoints is returned.
| bool smile::findPeriodicOrbit | ( | const CPotential * | potential, |
| double | Omega, | ||
| unsigned int | coord_section, | ||
| unsigned int | coord_first, | ||
| bool | threeD, | ||
| unsigned int | Nperiods, | ||
| CPosVelPoint< double > * | IC | ||
| ) |
Utility function to find a periodic orbit in the plane coord1:coord2 which is closest to the provided initial conditions.
| double smile::findRoot | ( | gsl_function * | F, |
| double | xlower, | ||
| double | xupper, | ||
| double | xdefault = 0, |
||
| double | reltoler = 1e-8 |
||
| ) |
wrapper for GSL root solver running multiple iterations until the convergence is achieved.
may additionally specify desired relative tolerance and default value to be returned if endpoints don't enclose root.
| bool smile::fitMassModel | ( | const std::vector< NumT > & | particlecoord, |
| const std::vector< NumT > & | particlevalue, | ||
| std::vector< double > * | gridcoord, | ||
| std::vector< double > * | gridvalue, | ||
| double | smoothing = 0, |
||
| size_t | gridsize = 0, |
||
| size_t | nskip = 0, |
||
| CMassModel::FITTYPE | ftype = CMassModel::FT_MASS, |
||
| const CMassModel * | model = NULL |
||
| ) |
Function that computes the input data for a spherical mass model from a given array of particles.
Depending on the parameter ftype, it may use the information on the radial mass profile, or on the distribution function in energy. There are three regimes:
model. | [in] | particlecoord | array of particle coordinates (if ftype==CMassModel::FT_MASS) or energies otherwise |
| [in] | particlevalue | array of particle masses |
| [out] | gridcoord | output grid in r, E, or h, depending on ftype |
| [out] | gridvalue | output the values of M(r) or f(E or h) |
| [in] | smoothing | smoothing parameter used in penalized spline smoothing |
| [in] | gridsize | size of the output grid, 0 means determine automatically |
| [in] | nskip | number of points to skip at the ends of grid |
| [in] | ftype | action to perform |
| [in] | model | existing mass model (if ftype!=CMassModel::FT_MASS) |
| const char * smile::getIntegratorNameByType | ( | CBasicOdeIntegrator::STEPPERKIND | integratorType | ) |
Correspondence between ODE integrator names and types return the name for a given integrator type
| const char * smile::getSchwDataNameByType | ( | CBasicSchwData::SCHWDATATYPE | dataType | ) |
Correspondence between Schw.model names and types.
return the name for a given Schwarzschild data type
| void smile::gsl_verbose_error_handler | ( | const char * | reason, |
| const char * | file, | ||
| int | line, | ||
| int | gsl_errno | ||
| ) |
error reporting routine which is invoked from inside GSL.
By default GSL crashes program on any error, which is not fun. To change this behaviour, add the following line at the start of the program: gsl_set_error_handler(&smile::gsl_verbose_error_handler);
| double smile::longAxisRadius | ( | const CPotential * | potential, |
| double | E, | ||
| double | Omega = 0 |
||
| ) |
Convenience function to find the maximal elongation of long-axis orbit of given energy.
(i.e. solve for Phi(x,0,0)=E ).
| void smile::my_stderr_show_message | ( | const std::string & | origin, |
| const std::string & | message | ||
| ) |
default routine that dumps text messages to stderr
default message printing routine
| void smile::readConfigOrbit | ( | const CConfigFile & | configReader, |
| CConfigOrbit * | ptrConfigOrbit | ||
| ) |
Read configuration data for orbit integration (a subset of parameters contained in the [Orbit] section of INI file).
| [in] | configReader | is the instance of class that handles the INI file; |
| [in,out] | ptrConfigOrbit | is the pointer to the configuration block that will be filled with the values read from the INI file; if no corresponding value exists in the file, then an old one is retained. |
| void smile::readConfigPotential | ( | const CConfigFile & | configReader, |
| CConfigPotential * | ptrConfigPotential, | ||
| unsigned int | potGroup = 0 |
||
| ) |
Read the configuration data from the INI file which is contained in one [Potential*] section.
| [in] | configReader | is the instance of class that handles the INI file; |
| [in,out] | ptrConfigPotential | is the pointer to the configuration block that will be filled with the values read from the INI file; if no corresponding value exists in the file, then an old one is retained. |
| [in] | potGroup | is the index of potential group (the first one is contained in the section [Potential], the second one in [Potential1], etc.). |
| const CPotential * smile::readPotential | ( | CConfigPotential * | configPotential | ) |
load a potential from a text or snapshot file.
The input file may contain one of the following kinds of data:
| [in,out] | configPotential | contains the potential parameters and may be updated upon reading the file (e.g. the number of expansion coefficients may change). If the file doesn't contain appropriate kind of potential (i.e. if configPotential->PotentialType is PT_NB but the file contains BSE coefficients or a description of MGE model), an error is returned. configPotential->NbodyFile contains the file name from which the data is loaded. |
| void smile::splitString | ( | const std::string & | src, |
| const std::string & | delim, | ||
| std::vector< std::string > * | result | ||
| ) |
routine that splits one line from a text file into several items.
| [in] | src | – string to be split |
| [in] | delim | – array of characters used as delimiters |
| [out] | result | – pointer to an existing array of strings in which the elements will be stored |
| double smile::totalEnergy | ( | const CPotential * | potential, |
| const CPosVelPoint< NumT > & | point, | ||
| double | Omega = 0, |
||
| double | t = 0 |
||
| ) |
Convenience function to return the total energy (or Jacobi integral if Omega!=0) for a given position+velocity point.
| void smile::tuneNonPrime | ( | size_t * | n | ) |
utility function to ensure that n does not have large prime divisors.
if needed, decrease it (in the FFT primes are great evil).
| void smile::writeConfigOrbit | ( | CConfigFile & | configWriter, |
| const CConfigOrbit * | ptrConfigOrbit | ||
| ) |
Write configuration data for orbit integration to INI file.
| [in,out] | configWriter | is the instance of class that handles the INI file (it will be updated); |
| [in] | ptrConfigOrbit | is the pointer to the configuration block for the orbit integration. |
| void smile::writeConfigPotential | ( | CConfigFile & | configWriter, |
| const CConfigPotential * | ptrConfigPotential, | ||
| unsigned int | potGroup = 0 |
||
| ) |
Write the configuration data for one potential component into the INI file.
| [in,out] | configWriter | is the instance of class that handles the INI file (it will be updated); |
| [in] | ptrConfigPotential | is the pointer to the configuration block for the potential component; |
| [in] | potGroup | is the index of potential section (0th is written to the section [Potential], 1st to section [Potential1], etc.) |
| bool smile::writePotential | ( | const std::string & | fileName, |
| const CPotential * | potential | ||
| ) |
write potential expansion coefficients to a text file.
The potential must be one of the following expansion classes: CPotentialBSE, CPotentialBSECompact, CPotentialSpline, CPotentialCylSpline, CPotentialScaleFreeSH. The coefficients stored in a file may be later loaded by readPotential() function.
| [in] | fileName | is the output file |
| [in] | potential | is the pointer to potential |
| const CConfigOrbit smile::defaultConfigOrbit |
default parameters for orbit integration and classification
| const CConfigPotential smile::defaultConfigPotential |
default parameters for all possible potentials
contains default potential parameters
| const double smile::FREQ_PRECISE_THRESHOLD =5.0 |
Use Hunter's method for precise determination of frequency of a given line if the spectrum neat its maximum is reasonably similar to that of a single line.
Otherwise use less precise method without Hanning filtering. Controls threshold for selection.
| const gsl_interp_type* smile::gsl_interp_ccspline |
a variant of cubic spline with given values of first derivative at endpoints.
these values are passed as two additional points in y[i] array ( y[npoints] for leftmost, y[npoints+1] for rightmost). if one wants to leave the standard, natural boundary condition (second derivative is zero), then pass NaN as a correspoinding 1st derivative parameter.
| const size_t smile::MAX_FFT_PRIME =41 |
orbit spectrum is calculated by fast fourier transform from GSL.
to avoid worst-case behavior of FFT, we ensure that its length never contains primes larger that MAX_FFT_PRIME (here equal to 42 minus unity)
| show_message_type * smile::my_message_ptr = &my_stderr_show_message |
global variable to the routine that shows information messages (emitted by various time-consuming routines to display progress), if NULL then no messaging is done
global variable to the routine that shows information messages.
called by various time-consuming routines to display progress, if NULL then no messaging is done
|
static |
 1.8.8
1.8.8